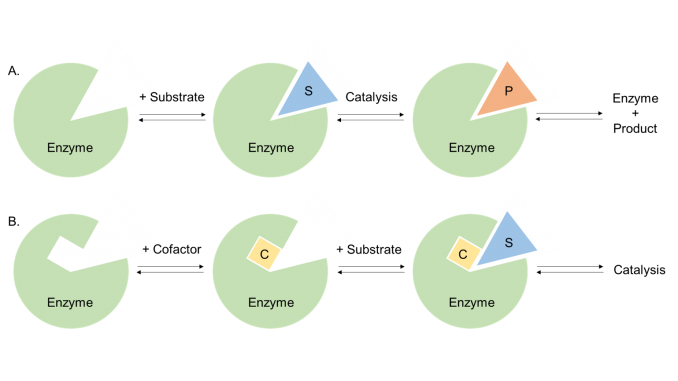
Figure 1. Schematic figure of enzyme plus substrate and cofactor.
All enzymes are proteins, but not all proteins are enzymes, the difference being that enzymes possess catalytic activity. The part of the enzyme tertiary structure which is responsible for the catalytic activity is called the ‘active site’ of the enzyme, and often makes up only 10–20% of the total volume of the enzyme. This is where the enzyme chemistry takes place. The active site is usually a hydrophilic cleft or cavity containing an array of amino acid side chains which bind the substrate and carry out the enzymatic reaction. In some cases the enzyme active site also binds one or more cofactors which assist in the catalysis of particular types of enzymatic reactions (Figure 1).
How does the enzyme bind the substrate? One of the hallmarks of enzyme catalysis is its high substrate selectivity, which is due to a series of highly specific non-covalent enzyme–substrate binding interactions. Since the active site is chiral, it is naturally able to bind one enantiomer of the substrate over the other, just as a hand fits a glove. There are four types of enzyme–substrate interactions used by enzymes, as follows:
(1) Electrostatic interactions. Substrates containing ionisable functional groups which are charged in aqueous solution at or near pH 7 are often bound via electrostatic interactions to oppositely charged amino acid side chains at the enzyme active site. Thus, for example, carboxylic acids are found as the negatively charged carboxylate anion at pH 7, and are often bound to positively charged side chains such as the protonated e-amino side chain of a lysine or the protonated guanidine side chain of arginine. Similarly, positively charged substrate groups can be bound electrostatically to negatively charged amino acid side chains of aspartate and glutamate.
(2) Hydrogen bonding. Hydrogen bonds can be formed between a hydrogen bond donor containing a lone pair of electrons and a hydrogen-bond acceptor containing an acidic hydrogen. These interactions are widely used for binding polar substrate functional groups. The strength of hydrogen bonds depends upon the chemical nature and the geometrical alignment of the interacting groups. Studies of enzymes in which hydrogen bonding groups have been specifically mutated has revealed that hydrogen bonds between uncharged donors/acceptors are of energy 2.0–7.5 kJ/mol, whilst hydrogen bonds between charged donors/acceptors are much stronger, in the range 12.5– 25 kJ/mol.
(3) Non-polar (Van der Waals) interactions. Van der Waals interactions arise from interatomic contacts between the substrate and the active site. Since the shape of the active site is usually highly complementary to the shape of the substrate, the sum of the enzyme–substrate Van der Waals interactions can be quite substantial (50–100 kJ/mol), even though each individual interaction is quite weak (6–8 kJ/mol).
(4) Hydrophobic interactions. If the substrate contains a hydrophobic group or surface, then favorable binding interactions can be realized if this is bound in a hydrophobic part of the enzyme active site. These hydrophobic interactions can be visualized in terms of the tendency for hydrophobic organic molecules to aggregate and extract into a non-polar solvent rather than remain in aqueous solution. These processes of aggregation and extraction are energetically favorable due to the maximization of inter-water hydrogen-bonding networks which are otherwise disrupted by the hydrophobic molecule.
There are many examples of hydrophobic ‘pockets’ or surfaces in enzyme active sites which interact favorably with hydrophobic groups or surfaces in the substrate and hence exclude water from the two hydrophobic surfaces. As mentioned above, these hydrophobic interactions may be very important for maintaining protein tertiary structure and they are central to the behavior of biological membranes. Having bound the substrate, the enzyme then proceeds to catalyze its specific chemical reaction using active site catalytic groups, and finally releases its product back into solution.
Reference:
Bugg T D H. Introduction to Enzyme and Coenzyme Chemistry. Natural Product Reports, 2005, 22(1):127-127.
Related Services at Creative Enzymes:

Leave a Reply